Professor Emeritus
Ph.D. Plant Pathology University of Wisconsin, Madison, 1977
B.S. Botany Oregon State University, 1973
![]() CV
CV
Our research group studies aspects of epiphytic bacteria that live on healthy plants' surfaces, emphasizing bacteria active in ice nucleation, causing frost damage to plants. We also study plant pathogenic bacteria that inhabit plant surfaces before infection. We use molecular genetic and ecological approaches to study how epiphytic bacteria interact with other microorganisms on plants, and with the plants on which they live. We seek to better understand adaptations epiphytic bacteria have evolved to exploit this unique habitat.
Molecular and ecological studies of plant-associated bacteria.
Our research group has focused on the molecular microbial ecology of plant-associated bacteria. While relatively un-studied, microorganisms that live on plant surfaces are enormously important in that this habitat serves as a reservoir of microbes that can infect the plants on which they live, can injure plants by acting as catalysts for ice formation (ice nucleation activity), and by producing plant hormones that can alter the normal pattern of plant development. As model systems I have focused much of this effort on the plant pathogenic bacterium Pseudomonas syringae, and the saprophytic bacterium Erwinia herbicola (Pantoea agglomerans) on plant surfaces. These are two of the bacteria that are most commonly found on plants. They offer the opportunity to compare and contrast the growth and survival tactics employed by pathogenic and non-pathogenic bacteria on the surface of healthy plants.

More recently the lab is also addressing the endophytic growth of bacteria within plants. In these studies we are comparing the strategies used by P. syringae, a facultative pathogen of plants that can multiply within the intercellular spaces of plant tissues without causing disease symptoms with that of Xylella fastidiosa, an obligate colonist of xylem vessels. To address these questions my program has developed and applied a variety of molecular genetic tools for the study of these bacteria while in their natural habitats in and on plants. Our studies of plant-associated bacteria have revealed that the interactions between the bacteria and the plant are, in fact, strongly conditioned by bacterial traits that were a result of their interaction with each other. We now have several major projects that address a variety of cell density-dependent traits in bacteria that mediate their growth and survival in and on plants. This exciting new research area promises to provide a new perspective on the way we visualize plant-microbe interactions and to provide new strategies to manage the diseases caused by plant pathogenic bacteria.
Studies of the nature of leaf surfaces
To better understand those traits bacteria require for growth and survival on leaves we have done extensive work to address the nature of the "microhabitats" in which bacteria live on leaves. We have shown that measures of such factors as carbon source abundance on a leaf as a whole are not predictive of the environment of bacteria at the very small scales at which they live. To address this we have developed new tools for the study of the expression of bacterial genes while bacteria reside in natural habitats such as on leaves or in the soil. I have shown that bacterial ice nucleation genes have properties that make them excellent reporters of the transcriptional activity of other genes to which they are fused. I thus have advanced the application of "reporter genes" to the study of microbial ecology in natural habitats.
For example, by fusing environmentally-responsive promoters to ice nucleation or gfp reporter genes in bacteria to produce "biosensors" we can assess the response of individual bacteria to its local environment. This work has led us to make the descriptions of the actual concentration of important nutrients such as ferric iron, fructose, ammonium ions, nitrate, and sucrose on plants at the scale of individual bacterial cells. Our recent use of use of modified gfp reporter genes with altered stability in cells, in particular, has provided unprecedented insight into the world of microbes in natural habitats. We are finding that a given habitat such as a leaf or root is extraordinarily diverse in the number of sites where resources such as sugars are available as well as the amounts of such resources at a site. In essence we find that a given root or leaf harbors thousands of individual "universes" where bacteria live in isolation of other microbes and where they compete only locally with one another. It is through understanding of such small-scale spatial processes that we should be able to progress toward understanding how we can change the normal microbial communities on leaves to avoid plant pathogenic bacteria and ice nucleation active bacteria that can be harmful to plants. Clearly, patterns of competition and secondary metabolite production are much more complex than others had anticipated.

Ice formation in citrus leaf
Several other such studies on the nature of plant habitats at small scales are underway. These will continue to be a major focus of the lab in the near future since they should provide useful information to better understand the natural world at the very small scale in which cells live in natural habitats.
Genes expressed on plants and affecting plant interactions
We have measured the in planta expression of bacterial genes that we expect to be important in interactions with plants as well as by identifying genes that are expressed only when they are on plants. For example, we have successfully identified a novel biosynthetic pathway for IAA production in Erwinia herbicola and have cloned most of the genes involved in this biosynthetic pathway. The cloning of these genes has made it possible for us to produce appropriate gene fusions to determine the activity of these genes in bacterial cells while they are on leaves; this has led to exciting results indicating that bacteria are triggered to produce plant-modifying chemical such as IAA by chemical and physical triggers from the plant itself.
A major thrust in the lab has been to identify genes in P. syringae cells that are expressed only when they are on a plant. To accomplish this we have developed a novel promoter-trapping strategy we term Habitat Inducible Rescue of Survival (HIRS), by which genes active on plants are fused to a promoterless locus that rescues a conditionally lethal phenotype in P. syringae while on leaves. This strategy has worked extraordinarily well, and we have identified over 135 loci that are selectively expressed on plants. The genes identified in this strategy are providing insight into the traits that coordinately are involved in adaptation to the plant habitat as well as complex anti-sense patterns of gene expression that apparently are involved in regulating gene expression on plants in many cases. This library of genes is a huge resource for future research that will address how these plant-inducible genes contribute to interactions of bacteria with healthy plants.

Bacterial brown spot lesions in bean leaf caused by Pseudomonas syringae
My lab has also been very active in pursuing the functional genomics of P. syringae. We have cooperated with DOE-JGI to obtain a completed genomic sequence of P. syringae strain B728a. Since this strain is a superb epiphyte as well as a plant pathogen, comparative genomics are being pursued to identify genes involved in epiphytic fitness by identifying genes not found in related strains such as P. syringae pv. tomato that are not good epiphytes. We already have identified several interesting genes that apparently confer novel means of interacting with the plant and with other organisms. For example, a gene in strain B728a has high homology to hetC, a gene that confers heterokaryon incompatability (a form of apoptosis) in many filamentous fungi. We are pursuing experimentation that addresses whether such a gene is a fungal-specific virulence factor that might enable P. syringae to kill fungal cells on plants to overcome nutrient limitation that they would otherwise face in this nutrient-poor environment. The results on this and other such projects suggested by examination of the genome of P. syringae are very promising, and such genomics studies will clearly be a thrust of the lab in the future.
Quorum sensing in bacteria on plants
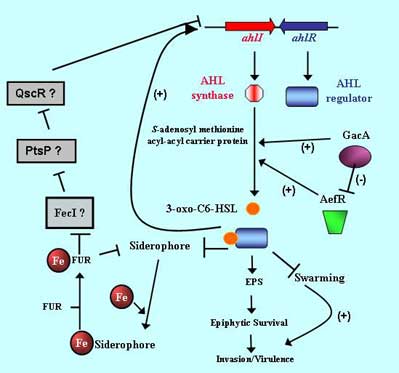
A major focus of the lab addresses how bacteria on plants benefit from cooperating with each other to ensure their survival or to interact with plants. We have found that a large percentage of the cells of pathogens such as P. syringae occur in aggregates on leaves. Through the use of Gfp-marked cells and viability stains we have found that such aggregates are required for tolerance of environmental stresses on leaves; while individual cells often die upon exposure to stressful conditions such as periodic desiccation on leaves, a high percentage of cells in aggregates survive such stresses. Furthermore, such bacterial aggregates facilitate the successful immigration of other cells to a leaf; immigrants of a variety of bacterial species that arrive at un- colonized sites have a much lower survival than if they had landed at a bacterial aggregate.
This work is leading to a new appreciation for the importance of aggregates in the biology of microbes on leaves, and has helped in our modeling efforts to describe the response of epiphytic populations to changes in their physical environment, and also to explain factors influencing successful immigration to a leaf. Such cell density-dependent behavior suggested that so-called quorum sensing may play an important role in the behavior of plant-associated bacteria. Most plant pathogenic bacteria produce small quorum-sensing signal molecules that act as co-inducers to regulate transcription of target genes in a cell density-dependent fashion. When the autoinducer of P. syringae, n-(3-oxo-hexanoyl)-L-homoserine lactone (AHL) is blocked by mutation, the epiphytic survival of mutants under stressful conditions on leaves was greatly reduced. Furthermore, we find that a variety of important traits in P. syringae such as swarming motility, extracellular polysaccharide production, oxidant tolerance, and certain virulence traits are altered in AHL mutants. Since the production of AHL quorum sensing molecules by this species is required for stress tolerance on leaves, the disruption of intercellular signaling on leaves could be an effective strategy of minimizing microbial colonization of leaves.
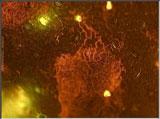
Large aggregate (more than 1000 cells) of cells of Pseudomonas syringae on bean leaf visualized after staining with acridine orange.
We are pursuing a genomics-based approach to determine what genes that are expressed in a cell density-dependent fashion in P. syringae and their coordinate regulation. Elucidation of the quorum sensing regulon should provide insight into other traits that are expressed in a cell density-dependent fashion, and that the contribution of those traits to the epiphytic and pathogenic behavior of P. syringae will be revealing. Since our work has already shown that cell-cell signaling is central to the process of infection of host plants by P. syringae, we are exploring how the infection process can be blocked by altering the abundance of AHL at the infection site. Our preliminary work has shown that the infection process can be blocked by supplying exogenous AHL in transgenic AHL-producing plants. We have several studies underway to test whether a "stealth model" of pathogenesis is common in different P. syringae pathovars and whether altering AHL content of plants might be a general new method of disease control.
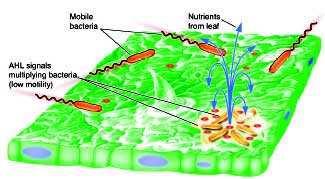
We have also found that the behavior of P. syringae on plants can be greatly affected by other epiphytic bacteria that produce AHL or that interfere with the ability of P. syringae to respond to its own AHL. This finding is an exciting new avenue of research in my lab that is producing evidence of cross talk among bacteria that leads to remarkable changes in behavior. Clearly, this finding leads to the prospect of new strategies of biological control of disease by "antagonistic" bacteria that alter AHL-mediated quorum sensing in P. syringae (and probably many other plant pathogens).
Studies of the endophytic plant pathogen Xylella fastidiosa
The study of cell-cell signaling in the endophytic bacterium Xylella fastidiosa recently became a major component of my research program. This bacterium is an important plant pathogen causing diseases of grape, and a variety of other important crop plants, but had been relatively unstudied due to its fastidious nature and lack of a genetic system. We have developed a tractable genetic system for X. fastidiosa and have made rapid progress toward understanding its colonization of xylem vessels of plants and its vectoring by insects. Our quantification of Gfp-marked strains of X. fastidiosa in plants reveal that, contrary to common perception, it spreads widely and forms modest sized aggregates on xylem vessels throughout the plant shortly after inoculation while blocking very few vessels by its growth. Such would be the pattern of colonization of vessels expected if it were principally growing as a biofilm- forming endophyte that would require xylem flow for its growth and maintenance in plants.

Pierce's disease of grape causing scorching of leaves and raisining of grape berries
Given that X. fastidiosa occurs as a classical "biofilm" in vessels and that examination of its genome content revealed the presence of a gene homologous to one in Xanthomonas campestris that conferred production of a small signal molecule (recently characterized as a fatty acid) that controls production of virulence factors such as cell wall degradation enzymes, we examined whether cell-cell signaling played a role in its colonization of plants. Mutants blocked in signal molecule production are hypervirulent to grape but do not colonize and hence are not transmitted by sharpshooter vectors. These findings further support our model that quorum sensing suppresses traits involved in biofilm formation in plants (to prevent deleterious effects of "over-colonization" of vessels) but promotes biofilm formation in insect vectors.
A great deal of work is underway in my lab to ascertain the regulon of the rpf cell-cell signaling locus and to determine other plant-inducible genes in X. fastidiosa by expression profiling using a DNA micro array we have developed for this species. Since our work suggests that the fatty acid signal molecule is required to suppress virulence of X. fastidiosa we are producing plants that express this signal molecule as well as exploring the production of signal molecule in plants by other endophytic bacteria as a means of altering the normal process of colonization by this plant pathogen. Our preliminary suggests that these strategies can achieve control of the important diseases caused by X. fastdiosa.
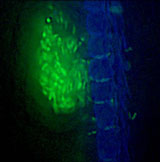
Movement of gfp-marked strains of Xylella fastidiosa across pit membranes of a grape xylem vessel
Applied Microbial Ecology
Because of my training and interest in Plant Pathology, I also have pursued research that addresses important problems related to plant productivity in California and worldwide. This work investigates important agricultural problems as model systems to develop concepts in microbial biology. I therefore have investigated in depth
- The epidemiology of an important bacterial disease of walnut trees caused by Xanthomonas campestris pv.juglandis
- The factors that can lead to successful biological control of fire blight, the most important bacterial disease of pear and apple worldwide caused by Erwinia amylovora
- New approaches by which frost injury to pear, potato, citrus and other important California crop plants caused by the ice nucleation active bacterium Pseudomonas syringae can be controlled
- The interactions of non-pathogenic bacteria such as Erwinia herbicola that incite fruit russeting of pear and apple by producing the plant hormone 3- indoleacetic acid (IAA).
A substantial amount of work in my laboratory is directed toward what might be called the "field ecology" of bacteria on plants. In this work we are addressing the population dynamics of bacterial populations on plants under field conditions and are investigating the factors that influence how many and what types of bacteria occur on plants and how they interact. Much of this work must be done in field studies, because the variable biological, physical and chemical environment on plants cannot be reproduced in laboratory and field conditions. In addition, many processes, such as immigration of bacteria to plant surfaces, occur at very large spatial scales.

Frost damage to potato plants following mild radiative frost of -4 C in field
The issues that we are addressing include the findings that:
- Bacterial populations on many plants such as citrus, and initial populations on deciduous tree crops is due primarily to immigration from nearby vegetation.
- Plant developmental abnormalities such as fruit russeting are associated with phytohormone-producing bacteria on plants. Alteration in the abundance and composition of nearby plant species and application of nitrogen-containing compounds that block IAA production in the bacteria can greatly reduce fruit russetting.
- Non-pathogenic, non ice nucleating bacteria such as Pseudomonas fluorescens strain A506 on trees, can inhibit the growth of other bacteria on plants by a process of pre-emptive competitive competition, leading to biological control of fire blight disease of pear and apple caused by Erwinia amylovora, as well as frost injury to plants caused by ice nucleating bacterial strains. This work has led to the commercial registration of this bacterium as a "biological pesticide" with the US EP A, enabling it to be used on many thousands of acres of high value crops for disease and frost control, resulting in large reductions in the use of antibiotics and other chemical pesticides that otherwise would have been used for control of these problems.
- Studies of the quantitative epidemiology of walnut blight disease caused by Xanthomonas arboricola pv. juglandis reveal that inoculum of the pathogen is found nearly exclusively in buds. This work has resulted in a model for the prediction of disease from estimates of pathogen inoculum abundance in buds in the winter, thus allowing disease control efforts to be applied only in orchards at risk of disease. The work has also revealed that early-season application of erradicant bactericides can yield excellent disease control while reducing the need for subsequent bactericide sprays, thereby reducing the cost of disease management and reducing environmental impacts of bactericide use.
- New methods of frost control for frost sensitive plants that I have developed depend on the assumption that ice nucleation caused by bacteria in a given plant part is the primary source of ice formation, and that ice propagation from distal plant parts on a plant is not important in the overall freezing process on a plant. Our on-going work has now shown that on plant species such as deciduous tree species (e.g. pear) that individual flowers freeze independently and that there is a large effect of reducing populations of ice nucleation active bacteria on frost susceptibility. On plants such as potato that have a dense canopy of overlapping leaves, ice propagation can be a significant source of ice formation and control of ice nucleating bacteria will have a lesser effect on frost damage.
1. Baccari, C., Killiny, N., Ionescu, M., Almeida, R.P.P., and Lindow, S.E. 2014. Diffusible signal factor-repressed extracellular traits enable attachment of Xylella fastidiosa to insect vectors and transmission. Phytopathology 104:27-33.
2. Roberts, E., and Lindow, S.E. 2014. Loline alkaloid production by fungal endophytes of Fescue species select for particular epiphytic bacterial microflora. ISME Journal 8:359-368.
3. Hockett, K.L., Burch, A.Y., and Lindow, S.E. 2013. Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. PLOS ONE 8: Article No.:e59850.
4. Lindow, S.E., Newman, K., Chatterjee, S., Baccari, C., Lavarone, A.T., and Ionescu, M. 2014. Production of Xylella fastidiosa diffusible signal factor in transgenic grape causes pathogen confusion and reduction in severity of Pierce’s disease. Molec. Plant-Microbe Interact. 27:244-254.
5. Hockett, K.L. Ionescu, M and Lindow, S.E. 2014. Involvement of rppH in thermo-regulation in Pseudomonas syringae. J. Bacteriol. 196:2314-2322.
6. Roper, C and Lindow S.E. 2013. Xylella fastidiosa: Insights into the lifestyle of a xylem-limited bacterium. Pp. 307-320 in: (N. Wang, J. Jones, G. Sundin, F. White, S. Hogenhout, C. Roper, L. De La Fuente, and J.H. Hams, eds.) Virulence mechanisms of plant pathogenic bacteria. APS Press. St. Paul, MN.
7. Retchless, A.C., Labroussaa, F., Shapiro, L., Stenger, D.C., Lindow, S.E. and Almeida, R.P.P. 2014. Genomic insights into Xylella fastidiosa interactions with plant and insect hosts. In: Genomics of plant-associated bacteria. (D. Gross, A. Lichens-Park and C. Kole, Eds). Springer.
8. Adams, R.I., Miletto, M., Lindow, S.E., Taylor, J. W., and Bruns, T.D. 2014. Airborne bacterial communities in residences: similarities and differences with fungi. PLOS ONE 9:issue 3 391283.
9. Burch, A.Y., Zeisler, V., Yokota, K., Schreiber, L., and Lindow, S.E. 2014. The hygroscopic biosurfactant syringafactin produced by Pseudomonas syringae enhances fitness on leaf surfaces during fluctuating humidity. Environ. Microbiol. 16:2086-2098.
10. Buchner, R.P., Gilles, C., Olson, W.H., Adaskaveg, J.E., Lindow, S. E., and Koutsoukis, R. 2014. Walnut blight management using Xanthomonas arboricola pv. juglandis dormant bud population sampling. Acta Hort. 1050:331-338.
11. Lindow, S.E., Olson, W., and Buchner, R.P. 2014. Colonization of dormant walnut buds by Xanthomonas arboricola pv. juglandis is predictive of subsequent disease. Phytopathology 104:1163-1174.
12. Caserta, R., Picchi, S.C., Takita, M.A., Tomaz, J.P., Pereira, W.E.L., Machado, M.A., Ionescu, M., Lindow, S.E., and de Souza, A.A. 2014. Expression of Xylella fastidiosa RpfF in citrus disrupts signaling in Xanthomonas citri subsp. citri and thereby its virulence. Molec. Plant-Microbe Interact. 27:1241-1252.
13. Ionescu, M., Zaini, P.A., Baccari, C., Tran, S., da Silva, A.M., and Lindow, S.E. 2014. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc. Natl. Acad. Sci. (USA). 111:E3910-E3918.
14. Yu, X., Lund, S.P., Greenwald, J.W., Records, A.H., Scott, R.A., Nettleton, D., Lindow, S.E., Gross, D.C., and Beattie, G.A. 2014. Transcriptional Analysis of the Global Regulatory Networks Active in Pseudomonas syringae during Leaf Colonization. mBio 5: doi:10.1128/mBio.01683-14.
15. Elkins, R.B., Temple, T.N., Shaffer, C.A., Ingels, C.A., Lindow, S.E., Zoller, B.G., and Johnson, K.B. 2015. Evaluation of dormant-stage inoculum sanitation as a component of a fire blight management program for fresh market Bartlett pear. Plant Disease 99:1147-1152.
16. Adams, R., Bhangar, S., Pasut, W., Arens, E.A., Taylor, J.W., Lindow, S.E., Nazaroff, W.W., and Bruns, T.D. 2015. Chamber bioaerosol study: Outdoor air and human occupants as sources of indoor airborne microbes. PLOS ONE. 10:e0128022.
17. Miletto, M. and Lindow, S.E. 2015. Relative and contextual contribution of different sources to the composition and abundance of indoor air bacteria in residences. Microbiome 3:61 DOI 10.1186/s40168-015-0128-z.
18. Scott, R.A., and Lindow, S.E. 2016. Transcriptional control of quorum sensing and associated metabolic interactions in Pseudomonas syringae strain B728a. Molec. Microbiol. 99:1080-1098.
19. Chatnaparat, T., Prathuangwong, S., and Lindow, S.E. 2016. Global pattern of gene expression of Xanthomonas axonopodis pv. glycines within soybean leaves. Molec. Plant-Microbe Interact. 29:508-522.
20. Lymperopoulou, D.S., Adams, R.I., and Lindow, S.E. 2016. Contribution of vegetation to the microbial composition of nearby outdoor air. Appl. Environ. Microbiol. 82:3822-3833.
21. Ionescu, M., Yokota, K., Antonova, E., Garcia, A., Beaulieu, E., Hayes, T., Iavarone, A.T., and Lindow, S. E. 2016. Promiscuous diffusible signal factor production and responsiveness of the Xylella fastidiosa rpf system. mBio 4:e01054-16.
22. Burch, A.Y., Do, P.T., Sbodio, A., Suslow, T.V., and Lindow, S.E. 2016. High culturability of epiphytic bacteria and frequency of biosurfactant producers on leaves. Appl. Environ. Microbiol. 82:5997-6009.
23. Gouran, H., Gillespie, H., Nascimento R., Chakraborthy, S., Zaini, P.A., Jacobson, A., Phinney, B.S., Dolan, D., Durbin-Johnson, B.P., Antonova, E.S., Lindow, S.E., Mellema, M.S., Goulart, L.R., and Dandekar, A.M. 2016. The secreted protease PrtA controls cell growth, biofilm formation, and pathogenicity in Xylella fastidiosa. Sci. Reports 6:31098.
24. Lymperopoulou, D.S., Coil, D.A., Schichnes, D., Lindow, S.E., Jospin, G., Eisen, J.A., and Adams, R.I. 2017. Draft genome sequences of eight bacteria isolated from the indoor environment: Staphylococcus capitis strain H36, S. capitis strain H65, S. cohnii strain H62, S. hominis strain H69, Microbacterium sp. strain H83, Mycobacterium iranicum strain H39, Plantibacter sp. strain H53, and Pseudomonas oryzihabitans strain H72. Standards in Genomic Sciences 12:17. DOI 10.1186/s40793-017-0223-9.
25. Lindow, S.E. 2017. Horizontal gene transfer gone wild: promiscuity in a kiwifruit pathogen leads to resistance to chemical control. Environ. Microbiol. 19:1363-1365.
26. Fabroussaa, F., Ionescu, M., Zeilinger, A.R., Lindow, S.E., and Almeida, R.P.P. 2017. A chitinase is required for Xylella fastidiosa colonization of its insect and plant hosts. Microbiology 163:502-509.
27. Caserta, R., Souza-Neto, R.R., Takita, M.A., Lindow, S.E., and De Souza, A.A. 2017. Ectopic expression of Xylella fastidiosa rpfF conferring production of diffusible signal factor in transgenic tobacco and citrus alters pathogen behavior and reduces disease severity. Molec. Plant-Microbe Interactions 30:866-875.
28. Adams, R.I., Lymperopoulou, D.S., Misztal, P.W., De Cassia-Pessotti, R., Behie, S.W., Tian, Y., Goldstein, A.H., Lindow, S.E., Nazaroff, W.W., Taylor, J.W., Traxler, M.F., and Bruns, T.D. 2017. Microbes and associated soluble and volatile chemicals on periodic wet household surfaces. Microbiome 5:128 DOI 10.1186/s40168-017-0347-6.
29. Zeilinger, A.R., Turek, D., Cornara, D., Sicard, A., Lndow, S.E., and Almeida, R.P.P. 2018. Bayesian vector transmission model detects conflicting interactions from transgenic disease-resistant grapevines. Ecosphere 9: Article No.: e02494.
30. Liu, Q., Lindow, S.E., and Zhang, J. 2018. Lactobacillus parafarranginis ZH1 producing anti-yeast substances to improve the aerobic stability of silage. Animal Sci. J. 89:1302-1309.
31. Miztal, P.K., Lymperopoulou, D.S., Adams, R.I., Scott, R.A., Lindow, S.E., Bruns, T., Tayler, J.W., Uehling, J., Bonito, G., Vilgalys, R., and Goldstein, A.H. 2018. Emission factors of microbial organic compounds from environmental bacteria and fungi. Environ. Sci. Technol. 52:8272-8282.
32. Baccari, C, Antonova, E., and Lindow, S. E., 2019. Biological control of Pierce’s disease of grape by an endophytic bacterium. Phytopathology 109:248-256.
33. Lindow, S. E. 2019. Money matters: Fueling recent insight into Xylella fastidiosa: an important and expanding global pathogen. Phytopathology 109:210-212.
34. Helmann, T. C., Deutschbauer, A. M., and Lindow, S.E. 2019. Genome-wide identification of Pseudomonas syringae genes required for fitness during the colonization of the leaf surface and apoplast. Proc. Natl. Acad. Sci. (USA) 116:18900-18910.
35. Helmann, T. C., Ongsarte, C. L., Lam, J., Deutschbauer, A. M., and Lindow, S. E. 2019. Genome-Wide Transposon Screen of a Pseudomonas syringae mexB Mutant Reveals the Substrates of Efflux Transporters. MBio 10:e02614-19.
36. Hernandez, M. N., and Lindow, S.E. 2019. Pseudomonas syringae increases water availability in leaf microenvironments by production of hygroscopic syringafactin. Appl. Environ. Microbiol. 85:e01014-19.
37. Czajkowski, R., Jackson, R. W., and Lindow, S. E. 2019. Editorial: Environmental bacteriophages: From biological control applications to directed bacterial evolution. Frontiers Microbiol. 10:1830.
38. Helmann, T. C., Deutschbauer, A. M., and Lindow, S.E. 2020. Distinctiveness of genes contributing to growth of Pseudomonas syringae in diverse plant species. PLOS ONE 15:e0239998.
39. Morella, N. M. Weng, F. C.-H., Joubert, P.M., Metcalf, C. J. E., Lindow, S. E. and Koskella, B. 2020. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc. Natl. Acad. Sci. (USA) 117:1148–1159.
40. Hernandez, M. N. and Lindow, S.E. 2021. Contact-dependent traits in Pseudomonas syringae B728a. PLOS ONE. 16:e0241655.
CNR Teaching Award - College of Natural Resources - 2004
Proctor and Gamble Award in Applied and Environmental Microbiology - American Academy of Microbiology - 2000
Member - National Academy of Sciences - 1999
Fellow - American Academy of Microbiology - 1999
Fellow - American Association for the Advancement of Science - 1999
Hildebrand-Laumeister Chair in Plant Pathology - College of Natural Resources - 1999
Alumnus of the Year - Oregon State University - 1995
Fellow - American Phytopathological Society - 1994
Ruth Allen Award - American Phytopathological Society - 1987
CIBA/GEIGY Award - American Phytopathological Society - 1985
Award for Initiatives in Research - National Academy of Sciences - 1985
Steven E. Lindow
Berkeley, California 94720