Associate Professor, Plant & Microbial Biology
Ph.D. Biochemistry, Molecular and Cell Biology, Johns Hopkins University School of Medicine, 1997
A.B. History of Science, Harvard College, 1990
The Ryan lab studies mechanisms such as phosphorylation, localization, and proteolysis that regulate protein activity in bacteria. Our model system is the Gram-negative freshwater bacterium Caulobacter crescentus, in which post-translational regulation plays an important role in cell cycle transitions, stress responses, antibiotic persistence, and the differentiation of one cell type into another. Ongoing projects in the lab include 1) the promotion of stress tolerance and antibiotic persistence by HipBA toxin-antitoxin systems, 2) the mechanisms which allow Caulobacter to survive without the essential outer membrane molecule lipid A, and 3) engineering Caulobacter strains for use in environmentally responsive living materials.
Research
Antibiotic persistence and stress resistance mediated by HipBA toxin-antitoxin systems
Unlike antibiotic resistance, which is caused by a heritable genetic change, persistence is a temporary dormant state in which individual bacteria can withstand doses of antibiotics that are otherwise lethal. Persistent sub-populations of bacteria can escape killing by antibiotics and later resume growth, causing recurrent infections.
We study how persistence is promoted in Caulobacter crescentus (a non-pathogen) by HipBA toxin-antitoxin systems. When the antitoxin HipB is depleted, free HipA toxin acts as a serine/threonine kinase to phosphorylate cellular proteins. Phosphorylation of HipA substrates can have several different effects, ranging from growth inhibition to stress resistance or antibiotic persistence.
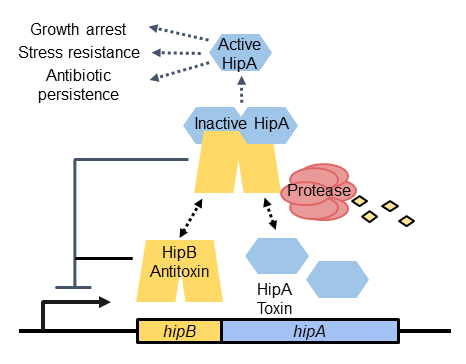
To study HipBA and persistence in Caulobacter, we are answering the following questions:
How are HipA toxins released from inhibition by HipB antitoxins?
What are the substrates of HipA toxins in Caulobacter?
How does phosphorylation affect the activity of each substrate?
Which substrates are important for promoting antibiotic persistence?
Structure, integrity, and remodeling of the outer membrane
The outer leaflet of the outer membrane (OM) of Gram-negative bacteria is composed chiefly of lipopolysaccharide (LPS), which provides a strong permeability barrier against harmful chemicals in the environment and is an important factor in the virulence of pathogens.
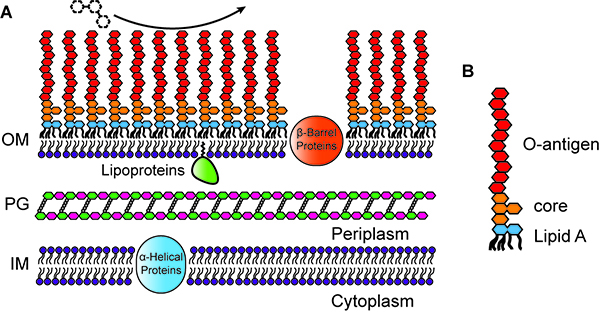
LPS itself has three parts: lipid A, a conserved core polysaccharide, and O-antigen, a highly variable polysaccharide. While the O-antigen and core polysaccharide are usually not required for bacterial viability, lipid A is nearly always essential for survival. Surprisingly, Caulobacter crescentus is viable in the absence of lipid A (and LPS) when the cells are grown in iron-limited medium, or when the iron-responsive transcriptional regulator Fur is inactivated. Caulobacter is only the fourth Gram-negative species shown to survive after elimination of its lipid A.
To understand the normal functions of lipid A and how Caulobacter adapts to its loss, we are asking the following questions:
1) Are specific Fur-regulated genes involved in survival after the elimination of lipid A?
2) What is the OM composed of when lipid A is absent?
3) Do stress response systems detect the absence of lipid A and promote survival and adaptation?
4) How do gene regulation and OM protein content change when lipid A is absent?
Lab Culture, Equity, and Inclusion
Selected Publications
Huang, C.Y., Gonzalez-Lopez, C., Henry, C., Mijakovic, I., Ryan, K.R. (2020) hipBA toxin-antitoxin systems mediate persistence in Caulobacter crescentus. Sci. Rep. 10: 2865.
Charrier, M., Li, D., Mann, V.R., Yun, L., Jani, S., Rad, B., Cohen, B.E., Ashby, P.D., Ryan, K.R., Ajo-Franklin, C.M. (2019) Engineering the S-layer of Caulobacter crescentus as a foundation for stable, high-density, 2D living materials. ACS Synth. Biol. 8: 181-190.
Smith, S.C., Joshi, K.K., Zik, J.J., Trinh, K., Kamajaya, A., Chien, P., Ryan, K.R. (2014) A cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc. Nat. Acad. Sci. USA 111: 14229-14234.
Shapland, E.B., Reisinger, S.J., Bajwa, A.K., and Ryan, K.R. (2011) An essential tyrosine phosphatase homolog regulates cell separation, outer membrane integrity, and morphology in Caulobacter crescentus. J. Bacteriol. 193: 4361-4370.
Ryan K.R., Taylor J.A., and Bowers L.M. (2010) The BAM complex subunit BamE (SmpA) is required for membrane integrity, stalk growth and normal levels of outer membrane ß-barrel proteins in Caulobacter crescentus. Microbiology 156:742-56.
Taylor J.A., Wilbur J.D., Smith S.C., and Ryan K.R. (2009) Mutations that alter RcdA surface residues decouple protein localization and CtrA proteolysis in Caulobacter crescentus. J. Mol. Biol. 394: 46-60.
Reisinger S.J., Huntwork S., Viollier P.H., and Ryan K.R. (2007) DivL performs critical cell cycle functions in Caulobacter crescentus independent of kinase activity. J. Bacteriol. 189: 8308-8320.
Biondi E.G., Reisinger S.J., Skerker J.M., Arif M., Perchuk B.S., Ryan K.R., and Laub MT. (2006) Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 444: 899-904.
McGrath P.T., Iniesta A.A., Ryan K.R., Shapiro L., McAdams HH. (2006) A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell 124:535-547.
Ryan K.R., Huntwork S., Shapiro L. (2004) Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc. Natl. Acad. Sci. USA. 101:7415-7420.
Ryan, K.R., Judd, E.M. and Shapiro, L. (2002) The CtrA response regulator essential for Caulobacter crescentus cell cycle progression requires a bipartite degradation signal for temporally controlled proteolysis. J. Mol. Biol. 324: 443-455.
Kathleen Ryan
Berkeley, CA 94720