Patricia C. Zambryski
Professor Emerita
Dr. Zambryski is a plant and microbial biologist and also serves as the department's Graduate Student Advisor
Ph.D. Molecular Biology University of Colorado, 1974
B.S. Genetics McGill University, 1969
PMB Head Graduate Student Advisor
Research
Agrobacterium and Plasmodesmata
Our laboratory performs research in two distinct areas. In microbial biology we study the molecular mechanisms utilized by Agrobacterium that leads to the genetic transformation of plant cells. In plant biology we study how plant cells communicate with each other via unique plant specific intercellular structures called plasmodesmata.
1. Agrobacterium mediated DNA transfer to plant cells. A specific DNA segment, T-DNA, is transferred from a large bacterial plasmid across bacterial and plant cell membranes and ultimately integrates stably into the plant nuclear genome. We study Agrobacterium-specific proteins and their respective molecular mechanisms responsible for producing a DNA-protein complex capable of plant cell transformation. Current research is aimed at defining the topology of the bacterial export channel. This channel is the paradigm for type IV secretion systems (T4SS). T4SS are utilized by plant and animal pathogens to transport DNA as well as protein toxins that manipulate their respective host cells to cause disease.
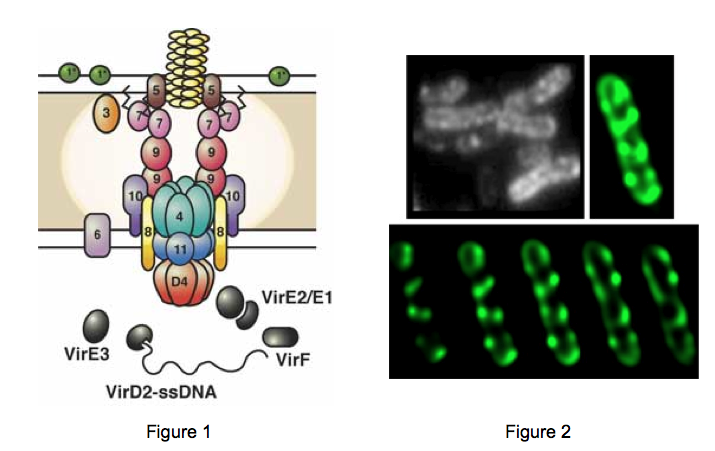
A model of how the 12 proteins (VirB1-B11, VirD4) essential for T4SS form a membrane-spanning channel is presented in Figure 1. The T-DNA is transferred as a single stranded intermediate called the T-strand. In addition, 4 proteins, VirD2, VirE2, VirE3, and VirF are exported by the T4SS to plant cells. This model was predicted by genetic and bioinformatics approaches. Current efforts aim to define the topology, location and function of the T4SS. We have recently determined that the multiple T4SS localize in a periodic (potentially) helical pattern around the circumference of the bacterial cell.This localization pattern was determined by deconvolution fluorescence microscopy of GFP fusions to T4SS components and substrates. Figure 2 shows several optical sections through the bacterial cell. This localization pattern was confirmed by immuno-fluorescence microscopy of native T4SS proteins. This localization pattern likely facilitates binding of Agrobacterium to susceptible plant cells. Indeed, if one tags the T4SS with GFP, one can see Agrobacterium binding laterally along their lengths to single plant cells. (Figure 2).
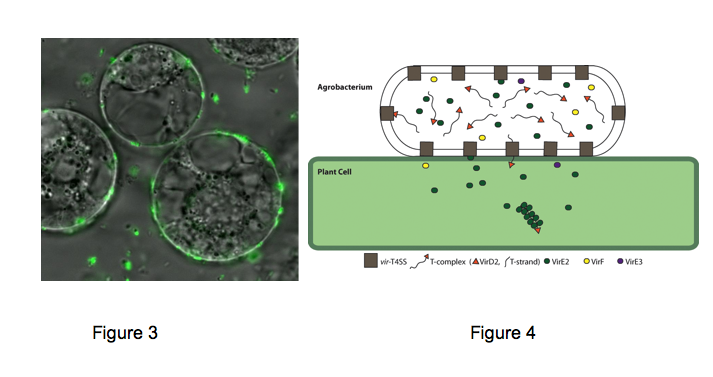
We now are investigating what constitutive bacterial components facilitate T4SS localization. We reasoned that the T4SS may either use an existing periodic/helical scaffold for its assembly, such as the bacterial cytoskeleton, or it may take advantage of interruptions in the bacterial cell wall (peptidogylcan) that occur during bacterial cell elongation. Recent studies suggest both hypotheses are correct and converge on the specific mechanisms underlying the determination of Agrobacterium cell shape. In addition we are determining the role of the extracellular T-pilus (colored in yellow Figure 1) in mediating plant cell contact and substrate transport.
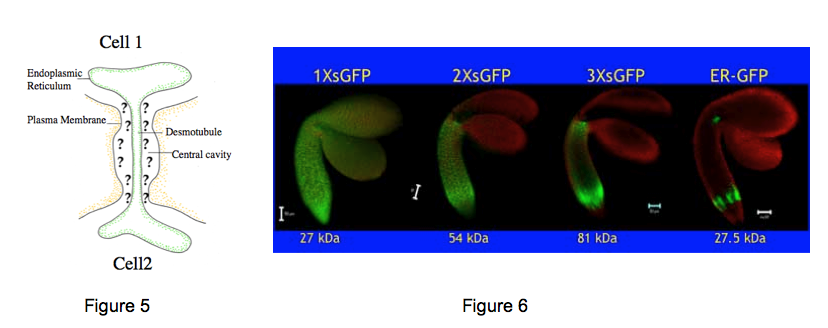
2. Plasmodesmata structure and function. Plasmodesmata (PD) are channels that span the plant cell wall, linking the cytoplasm between adjacent cells. These channels are highly dynamic, and impact on all levels of plant growth from general responses to the environment to defense against pathogens. Figure 5 shows that generic PD consist of membranes and spaces. The outer limits of PD are defined by the plasma membrane which is contiguous between two cells. The axial center of PD (called the desmotubule) is defined by endoplasmic reticulum between adjacent cells. The space between the desmotubule and the plasma membrane is called the cytoplasmic sleeve, and represents the contiguous cytoplasm between adjacent cells.
Our laboratory is interested in defining the factors that regulate PD structure and function that are localized at the entrance and along the length of the channel. We have developed a genetic screen using embryo defective lines of Arabidopsis and have several candidate mutants that define genes that affect PD aperture during mid embryogenesis.
We are also studying the innate capacity of plant embryos to traffic proteins cell to cell via PD. Different sized derivatives of green fluorescent protein (GFP) were initially expressed in meristematic regions of the embryo. Subsequent movement of encoded proteins from their site of synthesis then reveals PD aperture. 1X sized GFP moves throughout all regions of the embryo. 2X sized GFP cannot move to the embryonic leaves, and 3X sized GFP cannot move to the tip of the root. Thus, different regions of the embryo contain PD with different apertures that likely regulate the movement of critical regulators of development (Figure 6).
We are also studying the innate capacity of plant embryos to traffic proteins cell to cell via PD. Different sized derivatives of green fluorescent protein (GFP) were initially expressed in meristematic regions of the embryo. Subsequent movement of encoded proteins from their site of synthesis then reveals PD aperture. 1X sized GFP moves throughout all regions of the embryo. 2X sized GFP cannot move to the embryonic leaves, and 3X sized GFP cannot move to the tip of the root. Thus, different regions of the embryo contain PD with different apertures that likely regulate the movement of critical regulators of development (Figure 6).
We have identified the genes in two mutants with increased intercellular transport during embryogenesis, called increased size exclusion limit 1 (ise1) and ise2. ISE1 encodes a mitochondrial localized DEAD box RNA helicase, and ISE2 encodes a DEVH box RNA helicase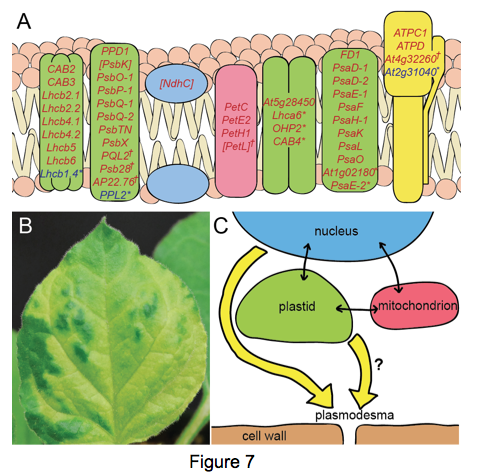 that localizes to chloroplasts. Whole genome expression analyses reveal that nuclear genes encoding plastid specific products are overwhelmingly down regulated in both ise1 and ise2 mutants (genes down regulated in both mutants are shown in red in Figure 7A). In fact, both mutants affect plastid development and cause leaf chlorosis (Figure 7B). These data imply extensive crosstalk among mitochondria, plastids, and nuclei. Since both mutants show increased de novo production of plasmodesmata, these expression analyses further imply that organelle-nuclear signaling affects plasmodesmata formation and function. While mitochondrial-nuclear or plastid-nuclear signaling are established and known retrograde signaling pathways, we have uncovered a novel pathway, dubbed organelle-nucleus-plasmodesmata signaling (ONPS)(Figure 7C). Independent analyses of redox states in mitochondria versus plastids confirm that organelle homeostasis affects plasmodesmata transport.Ongoing research aims to uncover genes and signals that regulate ONPS and de novo plasmodesmata synthesis.
that localizes to chloroplasts. Whole genome expression analyses reveal that nuclear genes encoding plastid specific products are overwhelmingly down regulated in both ise1 and ise2 mutants (genes down regulated in both mutants are shown in red in Figure 7A). In fact, both mutants affect plastid development and cause leaf chlorosis (Figure 7B). These data imply extensive crosstalk among mitochondria, plastids, and nuclei. Since both mutants show increased de novo production of plasmodesmata, these expression analyses further imply that organelle-nuclear signaling affects plasmodesmata formation and function. While mitochondrial-nuclear or plastid-nuclear signaling are established and known retrograde signaling pathways, we have uncovered a novel pathway, dubbed organelle-nucleus-plasmodesmata signaling (ONPS)(Figure 7C). Independent analyses of redox states in mitochondria versus plastids confirm that organelle homeostasis affects plasmodesmata transport.Ongoing research aims to uncover genes and signals that regulate ONPS and de novo plasmodesmata synthesis.
 that localizes to chloroplasts. Whole genome expression analyses reveal that nuclear genes encoding plastid specific products are overwhelmingly down regulated in both ise1 and ise2 mutants (genes down regulated in both mutants are shown in red in Figure 7A). In fact, both mutants affect plastid development and cause leaf chlorosis (Figure 7B). These data imply extensive crosstalk among mitochondria, plastids, and nuclei. Since both mutants show increased de novo production of plasmodesmata, these expression analyses further imply that organelle-nuclear signaling affects plasmodesmata formation and function. While mitochondrial-nuclear or plastid-nuclear signaling are established and known retrograde signaling pathways, we have uncovered a novel pathway, dubbed organelle-nucleus-plasmodesmata signaling (ONPS)(Figure 7C). Independent analyses of redox states in mitochondria versus plastids confirm that organelle homeostasis affects plasmodesmata transport.Ongoing research aims to uncover genes and signals that regulate ONPS and de novo plasmodesmata synthesis.
that localizes to chloroplasts. Whole genome expression analyses reveal that nuclear genes encoding plastid specific products are overwhelmingly down regulated in both ise1 and ise2 mutants (genes down regulated in both mutants are shown in red in Figure 7A). In fact, both mutants affect plastid development and cause leaf chlorosis (Figure 7B). These data imply extensive crosstalk among mitochondria, plastids, and nuclei. Since both mutants show increased de novo production of plasmodesmata, these expression analyses further imply that organelle-nuclear signaling affects plasmodesmata formation and function. While mitochondrial-nuclear or plastid-nuclear signaling are established and known retrograde signaling pathways, we have uncovered a novel pathway, dubbed organelle-nucleus-plasmodesmata signaling (ONPS)(Figure 7C). Independent analyses of redox states in mitochondria versus plastids confirm that organelle homeostasis affects plasmodesmata transport.Ongoing research aims to uncover genes and signals that regulate ONPS and de novo plasmodesmata synthesis.
Recent Publications
Agrobacterium mediated DNA transfer to plant cells
Aguilar, J., Cameron, T., Zupan, J., and Zambryski, P. Membrane and core periplasmic Agrobacterium tumefaciens virulence type IV secretion system components localize to multiple sites around the bacterial perimeter during lateral attachment to plant cells. mBio 2, in press. (2011)
Aguilar, J., Zupan, J., Cameron, T., and Zambryski, P. The Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence induced cells. Proc. Natl. Acad.Sci. USA, 107, 3758-3763 (2010)
Zupan, J., Hackworth, C., Ward, D., and Zambryski, P. VirB1* promotes T-pilus formation of the vir-type IV secretion system of Agrobacterium tumefaciens. J. Bacteriol., 189, 6551-6563 (2007)
Zupan, J., Hackworth, C., Ward, D., and Zambryski, P. VirB1* promotes T-pilus formation of the vir-type IV secretion system of Agrobacterium tumefaciens. J. Bacteriol., 189, 6551-6563 (2007)
Bailey, S., Ward., D., Middleton, R., Grossman, J.G., and Zambryski, P. Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc. Natl. Acad. Sci. USA 103 2582-2587 (2006)
Draper, O., Doucleff, M., Middleton, R., and Zambryski, P. Topology of VirB4 and VirB11 in the Agrobacterium tumefaciens VirB/D4 type IV secretion system. J. Biol. Chem. 281, 37628-35 (2006)
Middleton, R., Sjolander, K., Krishnamurthy, Foley, J., and Zambryski, P. Predicted hexameric structure of the Agrobacterium VirB4 C-terminus suggests VirB4 acts as a docking site during type IV secretion. Proc. Natl. Acad. Sci. USA 102, 1685-1690 (2005)
Middleton, R., Sjolander, K., Krishnamurthy, Foley, J., and Zambryski, P. Predicted hexameric structure of the Agrobacterium VirB4 C-terminus suggests VirB4 acts as a docking site during type IV secretion. Proc. Natl. Acad. Sci. USA 102, 1685-1690 (2005)
Plasmodesmata structure and function
Burch-Smith, T.M., and Zambryski, P. Plasmodesmata paradigm shift: regulation from without versus within. Ann. Rev. Plant Biol.Vol. 63, in press (2012)
Stonebloom, S., Brunkard, J., Cheung, A., Jiang, K., Feldman, L., and Zambryski, P. Redox states of plastids and mitochondria differentially regulate intercellular transport via plasmodesmata. Plant Physiol. In revision
Burch-Smith, T.M., Brunkard, J., Choi, Y.G., and Zambryski, P. Organelle-nucleus crosstalk regulates plant intercellular communication via plasmodesmata. PNAS Plus. In press (2011)
Burch-Smith, T., Stonebloom, S. Xu, M., and Zambryski, P. Plasmodesmata during development: re-examination of the importance of primary and secondary plasmodesmata structure versus function. Protoplasma 248, 61-74 (2011)
Burch-Smith, T., and Zambryski, P. Loss of INCREASED SIZE EXCLUSION LIMIT(ISE)1 or ISE2 increases the formation of secondary plasmodesmata. Curr. Biol. 20, 989-993 (2010)
Stonebloom, S., Burch-Smith,T., Kim, I., Meinke, D., Mindrinos, M., and Zambryski, P. Loss of the plant mitochondrial DEAD-box RNA helicase ISE1 leads to defective mitochondria and increased intercellular transport via plasmodesmata. Proc. Natl. Acad. Sci. USA 106, 17229-34 (2009)
Kobayashi, K., Otegui, M., Krishnakumar, S., Mindrinos, M., and Zambryski, P. INCREASED SIZE EXCLUSION LIMIT 2 encodes a DEVH box RNA helicase that regulates plasmodesmata during Arabidopsis embryogenesis. The Plant Cell 19, 1885- 1897 (2007)
Kim, I., Kobayashi, K, Cho, E., and Zambryski, P. Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 102, 11945-11950 (2005)
Kobayashi, K., Otegui, M., Krishnakumar, S., Mindrinos, M., and Zambryski, P. INCREASED SIZE EXCLUSION LIMIT 2 encodes a DEVH box RNA helicase that regulates plasmodesmata during Arabidopsis embryogenesis. The Plant Cell 19, 1885- 1897 (2007)
Kim, I., Kobayashi, K, Cho, E., and Zambryski, P. Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 102, 11945-11950 (2005)
Honors and Awards
AAAS (American Association for the Advancement of Science) Fellow, 2010
International Francqui Chair, University of Gent, Belgium, 2009 - Francqui Foundation - 2008
Miller Research Professorship - Miller Institute for Basic Research in Science - 2004
Fellow of the American Society for Microbiology - American Society for Microbiology - 2001
Member of the National Academy of Sciences - National Academy of Sciences - 2001
Miller Research Professorship - Miller Institute for Basic Research in Science - 2004
Fellow of the American Society for Microbiology - American Society for Microbiology - 2001
Member of the National Academy of Sciences - National Academy of Sciences - 2001
Patricia C. Zambryski
Research Focus:
Type IV secretion in bacteria and cell-to-cell transport in plants
E-mail
Location
281A Koshland
Berkeley, CA 94720
Berkeley, CA 94720
Phone Number
510.643.9203
Lab Phone Number
510.643.9204