Professor Emeritus

The Jackson Lab researched how viruses elicit plant diseases, and devised mechanisms for disease control in transgenic plants. We worked with three viruses: a plus sense monopartite RNA virus, tomato bushy stunt virus; a plus sense tripartite RNA virus, barley stripe mosaic virus; and a minus strand plant rhabdovirus, sonchus yellow net virus. We used genetic and biochemical analysis to investigate replication and movement of these viruses and to determine virus-host interactions culminating in disease.
Teaching
My major teaching commitment was Comparative Virology (a four unit upper division course) that I co-taught each Spring Semester since 1990. I also taught a Graduate Plant Virology (a three unit course), Co-Instructed Plant Biotechnology (a three unit upper division course), and offered a Freshman Seminar course, “Viruses, Health and Society” that enrolled a maximum of 15 talented entering freshman. For several years, I also have taught a six week Comparative Virology for graduate students at China Agricultural University in Beijing, and a one-month long graduate level Plant Virology minicourse at Zhejiang University in Hangzhou. I presented stimulating and contemporary lectures emphasizing scientific principles, encouraged interactive discussions, and provides my students with as much advice about career options as possible.
Plant viruses and disease control in transgenic plants
Our research goals were to understand how viruses elicit plant diseases and to devise mechanisms for disease control in transgenic plants. Two viruses with distinct strategies of replication emphasized in these studies. These include a negative strand plant rhabdovirus, sonchus yellow net virus (SYNV) that replicates in the nucleus, and a tripartite plus sense RNA virus, barley stripe mosaic virus (BSMV) that replicates in the cytoplasm.
Genetic and biochemical analyses were used to investigate the replication and movement of these viruses and to determine virus-host interactions culminating in disease. Each of the viruses engages in unique host associations during infection, and each virus gene has multiple roles in pathogenesis and mediates different host responses. Molecular genetic approaches and cytological studies used to follow infection processes are providing new information about the mechanisms of interactions with host cellular components during infection.
Plant rhabdoviruses
We had a major effort focused on studies of the plant rhabdoviruses. SYNV, the model system used for these studies, has a number of characteristics similar to those of rhabdoviruses that cause diseases of invertebrates and vertebrates. The rhabdoviruses have bacilliform (bullet-shaped) particles consisting of an envelope surrounding an internal nucleocapsid core that contains the negative sense genomic RNA. The animal rhabdoviruses replicate in the cytoplasm and the best studied example, vesicular stomatitis virus (VSV), encodes five proteins. However, SYNV differs from VSV by replicating in the nucleus and by encoding a sixth gene that is required for cell-to-cell movement of nucleocapsids. Three of the common proteins (designated N, P, and L) form the nucleocapsid core, which has an RNA-dependent-RNA-polymerase (RdRp) activity that functions in transcription of mRNAs and replication of the viral genomic and antigenomic RNAs. Virus particles also contain a matrix protein that coils the nucleocapsid and a surface glycoprotein that spans the viral envelope to associate with the matrix protein and the nucleocapsid core.
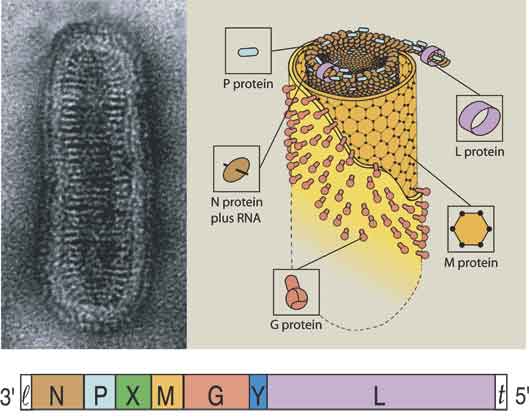
We showed that large subnuclear inclusions are formed during SYNV infection of plants and that these inclusions are the sites of viral replication. We isolated an RdRp complex from the nuclei of infected plants, and showed that the complex contains the N, P and L proteins plus the genomic RNA. Plant and yeast cells were used to characterize this complex by investigating the interactions of the virus and host proteins required for establishment of the subnuclear replication site. Our studies showed that homologous and heterologous interactions of the N and P proteins are required for formation of the subnuclear viroplasms, and that nuclear localization of the N and P proteins requires interactions with different nuclear import proteins.
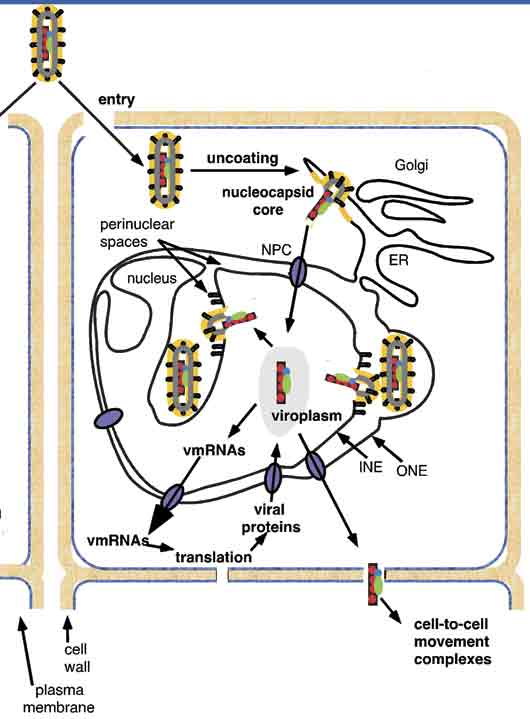
We also attempted to construct biologically active derivatives for genetic analyses of replication and pathogenesis. This is a challenging task because the nucleocapsid is the minimal unit of infectivity and must be assembled in cells in order to jumpstart replication.
Biological properties of the hordeiviruses
A second, long-term effort of the lab focused on the study of the biological properties of the hordeiviruses, exemplified by BSMV. Hordeivirus genomes consist of three positive sense RNAs. Using biologically active clones for genetic analyses, we showed that each of the seven genes encoded by the virus contribute to pathogenesis, and that the regulation of these genes and differences in their structure have profound effects on the disease phenotype in barley and other cereals, as well as Nicotiana benthamiana, and Chenopodium species.
Two distinct gene products encoded by RNAs α and ß are required for formation of the RdRp that associates with chloroplast outer membrane vesicles to form sites of viral replication. BSMV is distinct from many other viruses in that the coat protein is dispensable for the cell-to-cell and vascular movement functions. These functions require each of three proteins encoded in a triple gene block (TGB) on RNAß Our studies showed that the relative abundance of the TGB proteins is important for cell-to-cell movement and that the levels of the proteins are coordinated by transcription of two subgenomic messenger RNAs and unusual translational mechanisms.
Ectopic expression of various combinations of the TGB proteins were being used to probe their subcellular localization and to provide models for cell to cell movement of a nucleoprotein complex that can be recovered from infected plants. Pathogenicity functions of the hordeiviruses are mediated by a seventh protein, γb that is translated from a subgenomic mRNA derived from RNAγ. The γb protein has an N-terminal zinc finger and C-terminal coiled coil motifs whose mutations result in several distinctive disease phenotypes. RNA binding activities mediated by the zinc finger motif and protein-protein associations of γb are also critical for functions of γb that result in suppression of host silencing defenses.
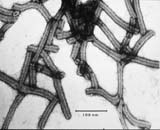
Electron micrograph of BSMV virions
Goodin, M.M., Dietzgen, R.G. Schichnes, D., Ruzin, S., and Jackson, A. O. (2002). pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31:375-383.
Johnson, J. A., Bragg, J. N., Lawrence, D. M., and Jackson, A. O. (2003). Mapping of the subgenomic RNA promoters of barley stripe mosaic virus. Virology 313:66-80.
Bragg, J. N., Lawrence, D. M., and Jackson, A. O. (2004) .The N-terminal 85 amino acids of the Barley stripe mosaic virus ϒb pathogenesis protein contain three zinc binding motifs. J. Virology 78:7379-7391.
Bragg, J. N., and Jackson, A. O. (2004). The C-terminal region of the Barley stripe mosaic virus ϒb protein participates in homologous interactions and is required for suppression of RNA silencing. Mol. Plant Pathol. 5:465-481.
Jackson, A. O., Dietzgen, R. G., Goodin, M. M., Bragg, J. N., and Deng, M. (2005). Biology of plant rhabdoviruses. Ann. Rev. Phytopathology 43:623-660.
Edwards, M. C., Bragg, J. N., and Jackson, A. O. (2006). Natural resistance mechanisms to viruses in barley. Ch. B8; pp 465-501. Natural Resistance Mechanisms of Plants to Viruses. Eds G. Lobenstein and J. P. Carr. Springer.
Deng, M., Bragg, J. N., Ruzin, S., Schichnes, D., King, D., Goodin, M., and Jackson, A. O. (2007). Role of the soncus yellow net virus N protein in formation of nuclear viroplasms. J. Virol. 81:5362-5374.
Lim, H-S., Bragg, J. N., Ganesan, U., Lawrence, D. M., Yu, J., Isogai, M., Hammond, J., and Jackson, A. O. (2008). Triple gene block protein interactions involved in movement of barley stripe mosaic virus. J. Virol. 82:4991-5006.
Jackson, A. O., Lim, H-S, Bragg, J. Ganesan, U., and Lee, M-Y. (2009). Hordeivirus replication, movement and pathogenesis. Ann. Rev. Phytopathology 47:385-422.
Renner, T., Bragg, J., Driscoll, H. E., Cho, J., Jackson, A. O. and Specht, C. D. (2009). Virus-induced gene silencing in the culinary ginger (Zingiber officinale): An effective mechanism for down-regulating gene expression in tropical monocots. Molecular Plant 1-11.
Lim, H-S, Bragg, J. N., Ganesan, U., Ruzin, S., Schichnes, D., Lee, M-Y., Varia, A. M., Ryu, K-H, Hammond, J., and A. O. Jackson (2009). Subcellular localization of the barley stripe mosaic virus triple gene block proteins. J. Virol. 83:9432-9448.
Verchot-Lubicz, J., Torrance, L., Solovyev, A. G., Morozov, S. Y., Jackson, A. O., and D. Gilmer (2010). Varied movement strategies employed by triple gene block encoding viruses. Mol. Plant-Microbe Interact. 23:1231-1247.
Covarrubias, S., Gaglia M., Kumar G. R., Wong W., Jackson, A. O., and B. A. Glaunsinger. 2011. Coordinated Destruction of Cellular Messages in Translation Complexes by the Gammaherpesvirus Host Shutoff Factor and the Mammalian Exonuclease Xrn1. PLoS Pathog 7(10): e1002339. doi:10.1371/journal.ppat.1002339.
Yuan, C., Li, C., Yan, L-J., Jackson, A. O., Liu, Z., Han, C., Yu, J., and D. Li. (2011). A high throughput Barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6(10): e26468. doi:10.1371/journal.pone.0026468.
Cui, Y., Lee, M-Y., Huo, N., Bragg, J. N., Yan, L., Yuan, C., Li, C., Holditch, S. J., Xie, J., Luo, M-C., Li, D., Yu, J., Martin, J., Schackwitz, W., Gu Y. Q., Vogel, J. P., Jackson, A. O., Liu, Z., and D. F. Garvin. (2012). Fine Mapping Barley Stripe Mosaic Virus Resistance Gene Bsr1 in the Model Grass Brachypodium distachyon. PLoS ONE 7(6): e38333. doi:10.1371/journal.pone.0038333.
Lee, M-Y., Yan, L., Gorter, F., Kim, B.Y-T., Cui, Y., Yuan, C., Grindheim, J., Ganesan, U. Liu, Z., Han C., Yu J., Li, D. and Jackson, A. O. (2012). A pathogenicity determinant overcoming Brachypodium distachyon Bd3-1 resistance resides in the Triple Gene Block 1 movement protein of the Norwich Strain of Barley stripe mosaic virus. J. Gen. Virol. 93:2729-2739.
Lim , H-S., Lee, M-Y., Moon, J. S., Moon, J-K., Yu, Y-M., Cho, I. S., Bae, H., Matt deBoer, M., Ju, H., Hammond, J., and Jackson, A. O. (2012). Actin cytoskeleton and Golgi involvement in Barley stripe mosaic virus movement and cell wall localization of triple gene block proteins. Plant Pathology 29:17-30.
Ganesan, U., Bragg, J. N., Deng, M., Marr, S., Lee, M-Y., Shi, M., Kappel, J., Peters, C., Lee, Y., Goodin, M. M., Dietzgen, R.G. and A. O. Jackson. (2013). Construction of a Sonchus Yellow Net Virus: A step toward reverse genetic analysis of plant negative strand viruses. J. Virol. 87:10598-10611.
Andrew O. Jackson
Ph.D. University of Manitoba, 1970