Associate Professor
Matt Traxler received his BS and PhD in microbiology from the University of Oklahoma,
and was a postdoctoral fellow at Harvard Medical School. His PhD studies used whole-genome
transcriptomics and network analyses to understand stress responses in E. coli.
His postdoctoral studies have included the use of NanoDESI and MALDI-TOF imaging
mass spectrometry to examine chemical exchange during actinomycete interactions.
His research aims include integrating metabolomic and transcriptomic paradigms with
the ultimate goal of understanding the role of specialized metabolism in bacterial
interactions and translating this knowledge into a platform for natural products discovery.
Most studies of bacteria are done in pure culture, investigating a single species in isolation. Yet, in natural environments, bacteria live in complex communities of interacting species.
My research explores the chemical ecology of interactions between actinomycete bacteria. Collectively, actinomycetes are the single deepest source of medicinal natural products, including antibiotics, antifungals, and anticancer agents (Fig. 1). Interactions between actinomycetes involve a remarkably rich chemical repertoire and provide a new approach for discovering novel natural products. My research program will incorporate whole-genome transcriptomics and newly developed mass spectrometry techniques to examine the physiology of actinomycete interactions as they relate to natural product biosynthesis and translate these insights into a new platform for natural products discovery.
Streptomyces coelicolor is the best-understood actinomycete. It is genetically tractable and its developmental lifecycle and secondary metabolism have been extensively studied. S. coelicolor makes multiple pigmented antibiotics whose production can be easily seen in colonies. These pigments provide a dramatic phenotype illustrating that co-cultivation alters the physiology of this bacterium.
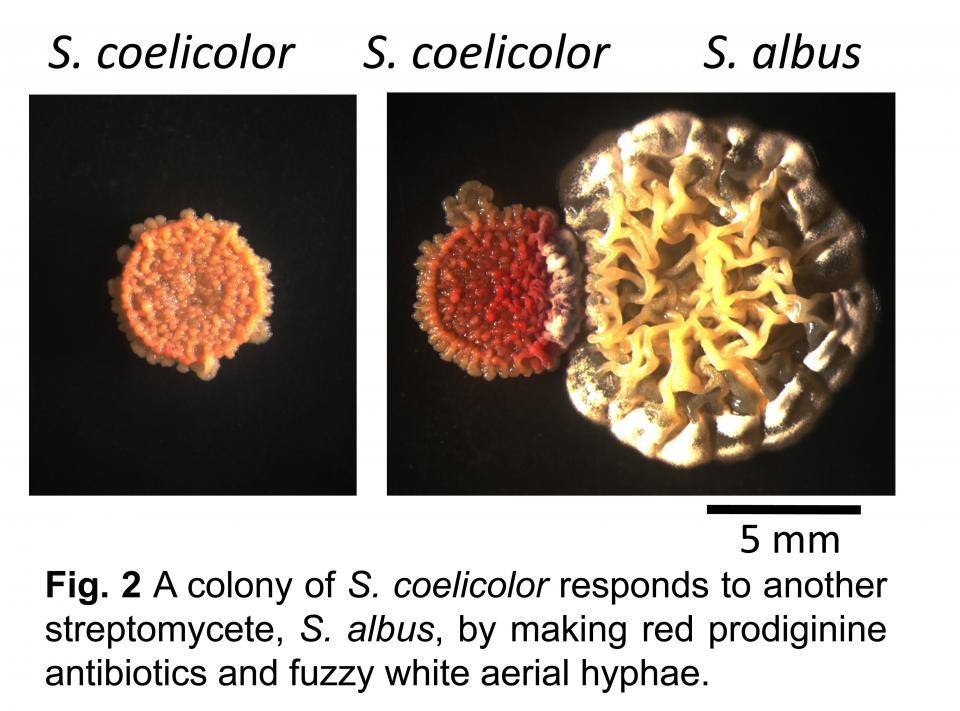
For example, the images in Fig. 2 show that co-cultivation with another actinomycete stimulates production of the red antibiotic prodigiosin, but this compound is not made in pure culture. However, this is merely the “tip of the iceberg” of the alterations in physiology that result from co-cultivation. My research uses innovative molecular biology, chemistry, and informatics techniques to understand the global changes that occur during actinomycete interactions and the role of specialized metabolites in actinomycete biology.
New tools for studying chemical exchange between bacteria
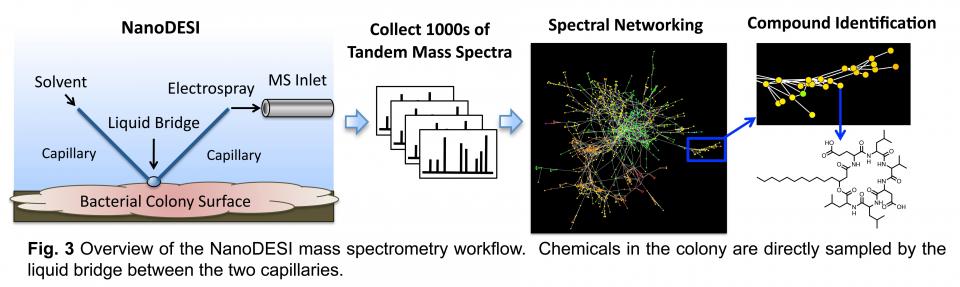
Acquiring deep and accurate chemical profiles of interspecies interactions as they occur is critical to understanding them. Two recently developed mass spectrometry techniques that have revolutionized the detection of molecules in bacterial colonies will be utilized in the Traxler lab. The first of these is nanospray desorption electrospray ionization mass spectrometry (NanoDESI MS). The NanoDESI MS workflow is shown schematically in Fig. 3. Compounds are desorbed directly from the sample surface in a small droplet of liquid. The compounds are then fragmented in the mass spectrometer, ultimately yielding individual tandem mass spectra for the thousands of ions detected. These metabolomic data are computationally assembled into networks that allow for systems-level interpretation and can guide compound identification. Thus, NanoDESI allows deep metabolomic sampling of a variety of biological surfaces.
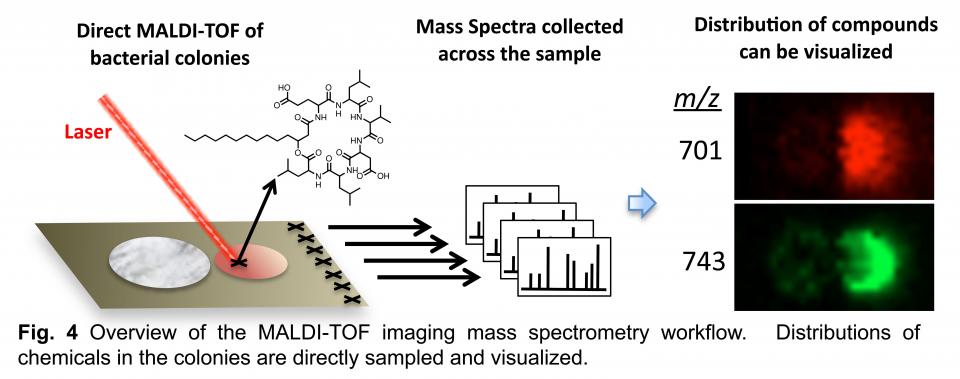
The second of these techniques is matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) imaging mass spectrometry (IMS) (Fig 4). It allows high-resolution mapping of compounds in microbial colonies. A laser is used to desorb molecules from the sample surface. Moving the laser in a grid format yields mass spectra from ordered points across the sample. These spectra are used to build images depicting the distribution of individual chemical species. This technique allows phenotypes to be correlated with the presence of chemical compounds.
An important aim is to couple mass spectral data with global transcriptional profiling of S. coelicolor to gain an in-depth view of the physiological changes triggered by interspecies interactions. This will entail obtaining chemical profiles and RNAseq data of S. coelicolor during interspecies interactions. These data will allow correlation of the expression of natural product biosynthetic genes with compounds observed in the samples. Subsequently, the expression of natural product biosynthetic genes can be connected with physiological responses also observed in the global transcriptome profiles.
The Traxler lab seeks to simultaneously build: 1) a framework for studying the physiology of interspecies interactions, and 2) an innovative program for natural products discovery. The unique combination of nascent metabolomic techniques and transcriptomics, complemented by a perspective rooted in bacterial physiology, will provide a much deeper picture of how interspecies interactions can impact specialized metabolism, and the role these compounds play in natural settings. The critical new information learned from these experiments will, in turn, be leveraged to maximum effect in the search for new natural products of potential therapeutic value.
Lambert S*, Traxler MF*, Craig M, Maciejewska M, Ongena M, van Wezel GP, Kolter R, and Rigali S. Altered desferrioxamine-mediated iron utilization is a common trait of bald mutants of Streptomyces coelicolor. Metallomics. 2014 Jul 23;6(8):1390-9.
Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, and Kolter R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. 2013 Aug 20;4(4).
Traxler MF, Kolter R. A massively spectacular view of the chemical lives of microbes. Proc Natl Acad Sci U S A. 2012 Jun 26;109(26):10128-9.
Seyedsayamdost MR*, Traxler MF*, Clardy J, Kolter R. Old meets new: using interspecies interactions to detect secondary metabolite production in actinomycetes. Methods Enzymol. 2012;517:89-109. PMID: 23084935
Traxler MF, Seyedsayamdost MR, Clardy J, Kolter R. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol Microbiol. 2012 Nov;86(3):628-44
Seyedsayamdost MR, Traxler MF, Zheng SL, Kolter R, Clardy J. Structure and biosynthesis of amychelin, an unusual mixed-ligand siderophore from Amycolatopsis sp. AA4. J Am Chem Soc. 2011 Aug 3;133(30):11434-7.
Romero D, Traxler MF, López D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011 Sep 14;111(9):5492-505. PMCID: PMC3173521
Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen HT, Stark SE, Conway T. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the 'feast to famine' gradient in Escherichia coli. Mol Microbiol. 2011 Feb;79(4):830-45
Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008 Jun;68(5):1128-48.
Traxler MF, Chang DE, and Conway T. Guanosine 3',5'-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc Natl Acad Sci U S A. 2006 Feb 14;103(7):2374-9.
Matt F. Traxler
Berkeley, California 94720-3102