Building mini magnets to navigate their environment
By Robert Sanders, UC Berkeley
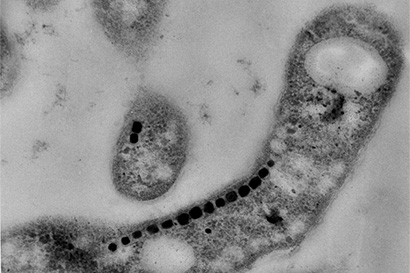
Many bacteria build miniature magnets and use them to navigate their environment and UC Berkeley’s Arash Komeili has found a neat trick they use to do it.
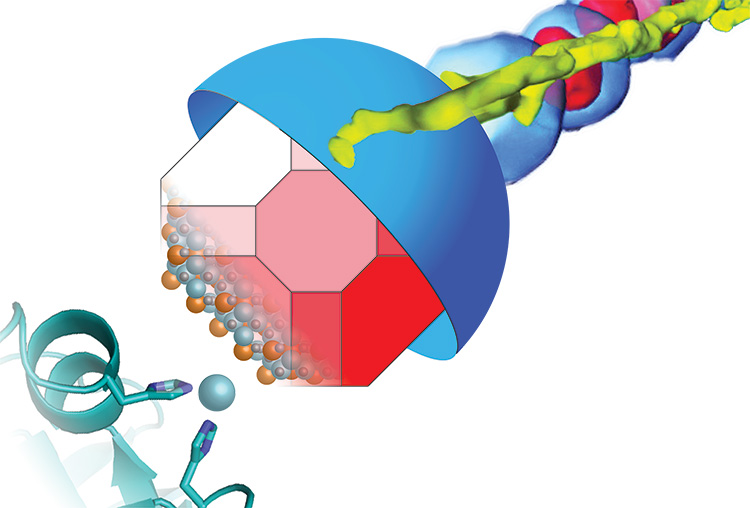
As reported in the March 2016 issue of the journal PLOS Biology, Komeili, an associate professor of plant and microbial biology, discovered that most so-called magnetotactic bacteria repurpose a commonplace enzyme and use it as a scaffold on which to assemble iron atoms into tiny magnets.
He and his colleagues discovered this repurposing of a deactivated enzyme in one species, Magnetospririllum magneticum, but when they looked throughout the three largest families of magnetotactic bacteria they found independent occurrences of the same story. A key feature of building magnetic particles is a run-of-the-mill protease – an enzyme that chews up proteins for recycling – that is deactivated.
“We really thought that something this unusual would have evolved only once. That just isn’t the case. It really just cements how unusual this process is,” said graduate student David Hershey.
Komeili is interested in how animals build minerals, a process known as biomineralization. Magnetotactic bacteria, for example, snatch iron atoms and assemble them into magnetic nanoparticles – tiny crystals of the mineral magnetite — that connect with their nervous system to help them navigate using Earth’s magnetic field as a reference. The new findings suggest that the scaffold protein, called MamO, may actually guide iron atoms into position in the magnetite crystal lattice.

“We would like to know how minerals are built in nature since they constitute a fundamental survival strategy for many organisms,” he said.
Their finding, based on X-ray crystallographic images of the MamO protein obtained through a collaboration with James Hurley’s lab, demonstrates once again that evolution uses existing molecules, such as ubiquitous proteases, for new functions, in this case a magnetic scaffold.
It also opens the door to using bacteria’s tricks to build unique magnetic minerals in a test tube that are not possible to build with normal chemistry.
 Komeili’s UC Berkeley co-authors include graduate student Patrick Browne, postdoctoral fellows Ertan Ozyamak and Stephanie Jones, research scientist Xuefeng Ren, Michelle Chang, an associate professor of chemistry, and James Hurley, a professor of molecular and cell biology. Ryan Melnyk of Lawrence Berkeley National Laboratory is also a co-author.
Komeili’s UC Berkeley co-authors include graduate student Patrick Browne, postdoctoral fellows Ertan Ozyamak and Stephanie Jones, research scientist Xuefeng Ren, Michelle Chang, an associate professor of chemistry, and James Hurley, a professor of molecular and cell biology. Ryan Melnyk of Lawrence Berkeley National Laboratory is also a co-author.
Komeili also has an appointment in the Department of Molecular and Cell Biology and is an affiliate of the California Institute for Quantitative Biosciences.
Links
To learn more, read the research summary by PLOS Biology.
MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria (PLOS Biology)